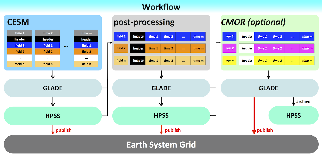
Very General CESM Workflow:
Current Workflow:
- Enter proposed experiment in the Experiment DB for approvals
- Configure and run
- Generate interum diagnostics
- Archive model output (short and long term)
- Review Diagnostics
- Post-process (aka "fast orange") to generate time-series data per variable(s) from raw history file output
- ssh [post-proccessing-host]
- in ~$HOME run "svn export https://subversion.ucar.edu/CCP_Processing_Suite" --- this uses the subversion source code control to build the processing suite locally remotely run "svn export https://proxy.subversion.ucar.edu/CCP_Processing_Suite"
- cd CCP_Processing_Suite
- Process_Setup [$CASE] [$HIST] [$TPER] [$PROC] [$CMIP] example: ./Process_Setup b40.rcp8_5.ldeg.001 clm2.h0 mon 1 0 --- this is a script the reads the experiments.txt file to match the 1st argument [CASE]. Can also run with no args to see list of expected args
- comes back with "./b40.rcp8_5.ldeg.001_clm2.h0_process.sh ready to run"
- nohup ./b40.rcp8_5.ldeg.001_clm2.h0_process.sh >& log_rcp8_5.1deg.clm2.h0 --- this starts the process takes a long time and builds *lots* of files. It runs in the background and sends stderr, stdout to the log file log_rcp8_5.1deg.clm2.h0. Note: the naming convention of the logfile should follow log_[$CASE].[$HIST].[$TPER]
- cd /datalocal/ccpg/aliceb/b40.rcp8_5.ldeg.001/lnd/mon to see netCDF files being built
- after run is complete, can cd /datalocal/ccpg/aliceb and remove all files and dir with rm -rf /datalocal/ccpg/aliceb/b40.rcp8_5.ldeg.001
- Write up scientific results
- Publish output to ESG or other archive sources (web, etc..)
Proposed Workflow for CESM1.3 and higher
- Enter proposed experiment in the Experiment DB for approvals
- Configure and run - PIO generates time-series output files as part of the model run
- Generate interum diagnostics
- Archive model output and rundb metadata (short and long term - compression may take place here...)
- Review Diagnostics
- Write up scientific results
- Publish output to ESG or other archive sources (web, etc..)
Proposed Workflow for CESM1.0.X, CESM1.1.X and CESM1.2.X
- Enter proposed experiment in the Experiment DB for approvals
- Configure and run
- Use new "fast orange" offline utility being generated as part of this project (converts to time series - and possibly compression?)
- Archive model output and rundb metadata (short and long term - compression may take place here...)
- Review Diagnostics
- Write up scientific results
- Publish output to ESG or other archive sources (web, etc..)
Completed Task List - updated 10/22/2013
- Set up Trello for managing tasks
- Change long term archiver to have an option to run without deleting files (Andy)
- Add two env_run.xml variables, one logical to activate (DOUT_S_SAVE_ALL_ON_DISK) and another optional one to specify the save directory (DOUT_S_SAVE_ROOT)
- These two env vars are accessed in lt_archive.sh; the default value for DOUT_S_SAVE_ROOT is $DOUT_S_ROOT with "archive" replaced with "csm"
- If activated, $DOUT_S_ROOT is fed to the "cp -al" command; this makes a copy of the directory tree in $DOUT_S_SAVE_ROOT; the leaves (files) are hard links
- Status: Done - changes to lt_archive.sh and env_run.xml
- Ongoing Community communication
- Evaluation by scientists if this new workflow fulfills their requirements
- UV-CDAT from DOE - How does this fit in (or not)?
- Internat'l community on-board with workflow changes for CMIP6 so need to communicate our work with them and vis-a-vis (e.g. Max Plank DB mgmt integration with workflow, Hadley using CMORE directly, etc...)
Some relevant documents:
CESM Experiment Case Name Convention (May 2011)
CCSM3 Output Filename Requirements (Jun 2005)
Here's a thumbnail of a diagram that I created for my CESM 2013 Workshop poster that show the workflow. It's relatively abstract.
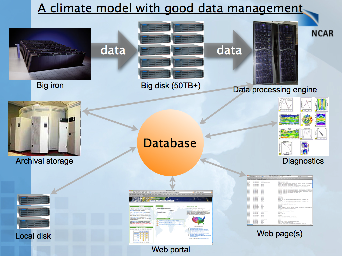
Here are two slides from a presentation I gave at the 2005 CISL User Forum that illustrate some basic ideas I had at the time:
A table of possible model output streams
component | stream name | convention | typical? |
|---|---|---|---|
atm | cam2.h0 | monthly averages | always |
atm | cam2.h1 | daily | almost always |
atm | cam2.h2 | daily/subdaily | sometimes |
atm | cam2.h3 | daily/subdaily | uncommon |
atm | cam2.h4 | daily/subdaily | rare |
atm | cam2.h5 | daily/subdaily | very rare |
atm | cam.h0 | monthly averages | always |
atm | cam.h1 | daily | almost always |
atm | cam.h2 | daily/subdaily | common |
atm | cam.h3 | daily/subdaily | less uncommon |
atm | cam.h4 | daily/subdaily | very rare |
atm | cam.h5 | daily/subdaily | none yet |
lnd | clm2.h0 | monthly averages | always |
lnd | clm2.h1 | daily | sometimes |
lnd | clm2.h2 | daily/subdaily | rare |
ice | cice.h | monthly averages | always |
ice | cice.h1 | daily | sometimes |
ice | cice.h2 | daily/subdaily | rare |
ice | cice.h2_06h | 6-hourly | CESM1-CAM5-BGC LE only |
ocn | pop.h | monthly averages | always |
ocn | pop.h2 | daily | uncommon |
ocn | pop.h.nday1 | daily | almost always |
ocn | pop.h.ecosys.nday1 | daily | ocean BGC only, common |
ocn | pop.h.ecosys.nyear1 | annual | ocean BGC only, common |
rtm | rtm.h0 | monthly averages | always |
rtm | rtm.h1 | daily | often |